
īinning: BI-FastQC uses a customizable binning strategy with a default of 5bp bins, while DRAGEN uses an algorithmic binning strategy based on the Granularity setting resulting in DRAGEN providing more precise results at the default settings.The most significant differences are described below. There are some differences in how the values are calculated and the resulting metrics will not be identical between the two tools. The reimplementation of FastQC in DRAGEN has been modified to take advantage of the hardware-acceleration provided by DRAGEN's Field-Programmable Gate Array (FPGA) for a significant speed improvement. To use this option, enter -fastqc-only=true to the DRAGEN command.ĭRAGEN's FastQC module is a complete reimplementation of the original FastQC tool developed by the Babraham Institute (BI-FastQC). The workflow only outputs key metric files and runs ~70% faster. If DNA alignment is not needed, or if QC results are needed more quickly, the mapping and BAM output portions of the workflow can be disabled. If you are only interested in sample QC or would like to obtain FastQC results only, DRAGEN provides a mode to generate the fastqc_metrics.csv file directly.īy default DRAGEN FastQC and read-trimming are run as preprocessing steps to standard sequence alignment workflows. All metrics are calculated and reported separately for each mate-pair. The metrics are generated automatically on all DRAGEN map-align workflows, with no additional run time, and output in a CSV format file called.
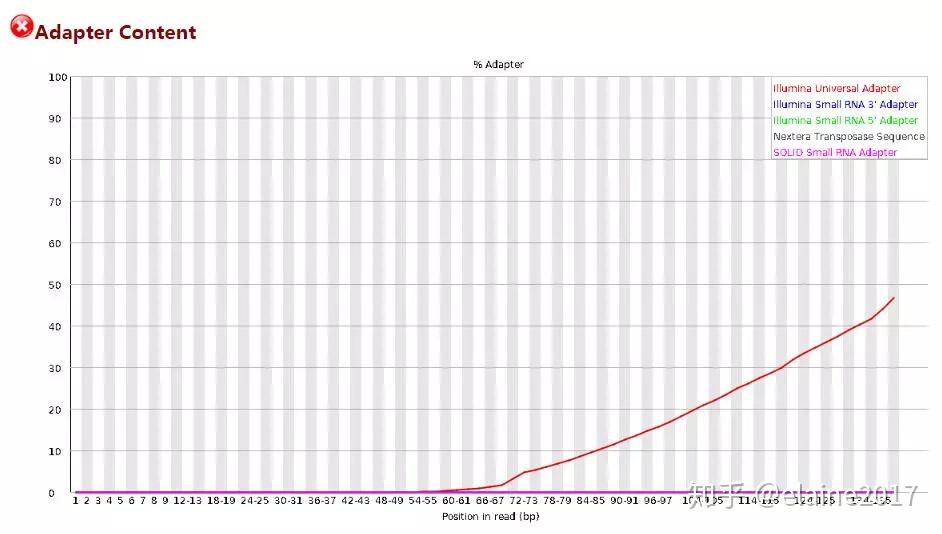
The tool is modeled after the metrics generated by the FastQC tool from Babraham Institute. The DRAGEN FastQC module is a tool for calculating common metrics used for quality control of high-throughput sequencing data.


 0 kommentar(er)
0 kommentar(er)
